|
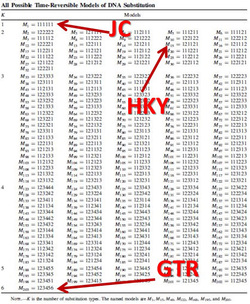
nst=mixed. This is one of my favorite commands in MrBayes. It's used instead of designating a nucleotide substitution model a priori. Instead, reversible jump is a form of model-averaging (across different dimensions of parameter space) where all possible time-reversible substitution models are explored, incorporating uncertainty in model selection.
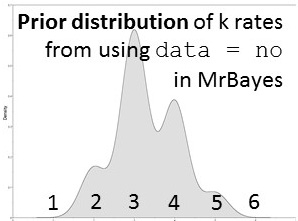
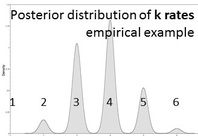
Likelihood-based analyses results are dependent on the model-fit to the data. The credible set of substitution models typically contains several different models and the benefit of rjMCMC is that it allows integration of all. Though tree topology isn't extremely sensitive to model misspecification, other parameters may be (Alfaro & Huelsenbeck 2006). example of mrbayes block using rjMCMC: [after the nexus alignment of data, use these commands for rjMCMC in MrBayes; this shows data grouped in two subsets] BEGIN MRBAYES; log start filename=murray-rj_log; charset codon_pos1 = 1-1041\3; charset codon_pos2 = 2-1041\3; charset codon_pos3 = 3-1041\3; partition two_subsets = 2: codon_pos1 codon_pos2, codon_pos3; set partition = two_subsets; lset applyto=(all) nst=mixed rates=gamma; [each of the two subsets will be treated separately] [use 'nst= mixed' for rjMCMC and 'rates=gamma' to incorporate rate heterogeneity; I don't use parameter for invariant sites] unlink shape=(all) pinvar=(all) statefreq=(all) revmat=(all); prset applyto=(all) ratepr=variable; mcmc ngen=1000000 samplefreq=100 filename=murray_rj; sumt; sump; [gives a default 25% burnin for tree and parameter files] log stop; end; Also a great tool -- rjMCMC in BEAST 2! Just install the RBS plugin while in BEAUti and you are set to go. The drawback -- as of now (Dec. 2016) you cannot run the XML file in CIPRES if you are using reversible jump models from the RBS plugin. note:
references:
Huelsenbeck, J.P., Larget, B. & Alfaro, M.E. (2004) Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol, 21, 1123-33. Alfaro, M.E., & Huelsenbeck, J.P. (2006). Comparative performance of Bayesian and AIC-based measures of phylogenetic model uncertainty. Syst Biol, 55, 89-96.
0 Comments
|
PhyloBlogCovering topics of phylogenetics and systematics & other science-related news. Archives
October 2019
Categories
All
|
 RSS Feed
RSS Feed