|
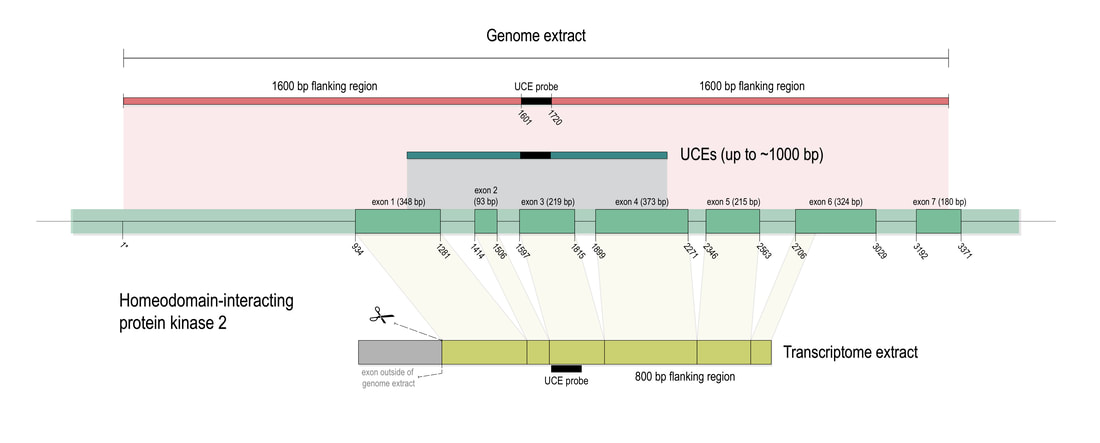
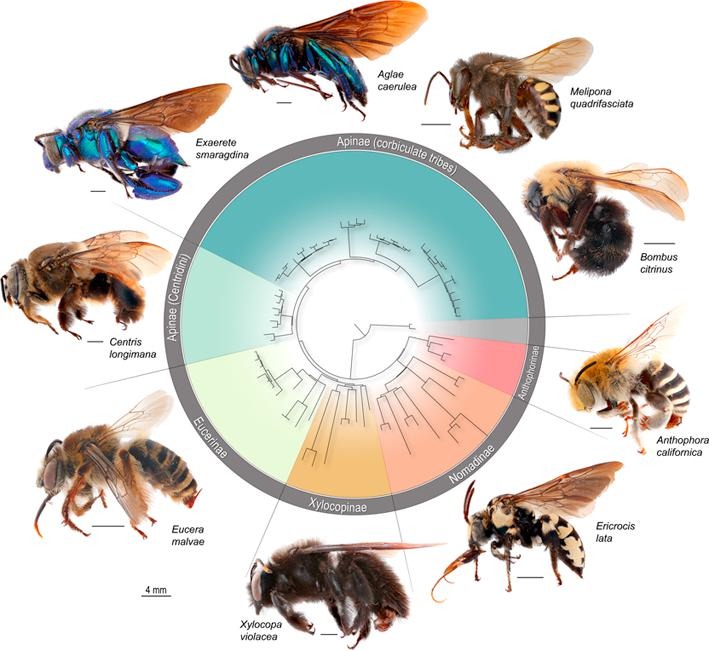
Phylogenomic methods are incredibly popular, and there are various baits and probes for capturing different pieces of the insect genome for analysis. One of the next aspects of 'big data' will be to develop ways to combine all of these sources! My previous post covered a method for visualizing data on tree tips -- specifically I showed a plot of the number of loci recovered after doing a quick combination of genomes, transcriptomes, and UCEs (using the Phyluce pipeline). I'm happy to write that our research paper on the combination of phylogenomic data has been published in Molecular Phylogenetics and Evolution. Bossert et al. 2019 report successful results when three types of datasets are combined for Apidae (the largest bee family).
references:
Bossert, S., Murray, E.A., Almeida, E.A.B., Brady, S.G., Blaimer, B.B. & Danforth, B.N. (2019) Combining transcriptomes and ultraconserved elements to illuminate the phylogeny of Apidae. Molecular Phylogenetics and Evolution, 130, 121-131. doi.org/10.1016/j.ympev.2018.10.012 Kieran, T.J., Gordon, E.R., Forthman, M., Hoey-Chamberlain, R., Kimball, R.T., Faircloth, B.C., Weirauch, C. and Glenn, T.C. (2019) Insight from an ultraconserved element bait set designed for Hemipteran phylogenetics integrated with genomic resources. Molecular Phylogenetics and Evolution, 130, 297-303.
0 Comments
Your comment will be posted after it is approved.
Leave a Reply. |
PhyloBlogCovering topics of phylogenetics and systematics & other science-related news. Archives
October 2019
Categories
All
|
 RSS Feed
RSS Feed