|
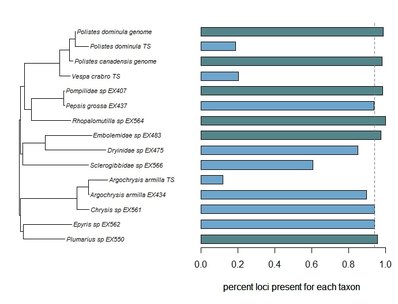
Phylogenomic datasets can be gappier than matrices concatenated from a small number of hand-selected, Sanger-sequenced genes. Here is a nice way to visualize the percent data coverage and see the distribution of missing data across the tree. I have a dataset of hundreds of loci acquired using ultraconserved element (UCE) probe matching. Besides my own data, there are other types of data that can potentially be incorporated (published genomes, transcriptomes, etc.). I want to see how the topology may be sensitive to different data and parameter permutations. I can use this handy barplot visual for dataset exploration, such as identifying problem clades with low data coverage.
Programs and packages involved in producing this figure:
Load your tree and your data (count data, continuous measurements, etc.) into R, use the handy script from Liam Revell's site, and in short time, you'll have a beautiful and informative figure! references:
Branstetter, M.G., Danforth, B.N., Pitts, J.P., Faircloth, B.C., Ward, P.S., Buffington, M.L., Gates, M.W., Kula, R.R. & Brady, S.G. (2017) Phylogenomic Insights into the evolution of stinging wasps and the origins of ants and bees. Current Biology, 27, 1019-1025. Branstetter, M.G., Longino, J.T., Ward, P.S. & Faircloth, B.C. (2017) Enriching the ant tree of life: enhanced UCE bait set for genome-scale phylogenetics of ants and other Hymenoptera. Methods in Ecology and Evolution, 8, 768-76. Faircloth, B.C. (2015) PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics, btv646. Lopez-Osorio, F., Pickett, K.M., Carpenter, J.M., Ballif, B.A. & Agnarsson, I. (2017) Phylogenomic analysis of yellowjackets and hornets (Hymenoptera: Vespidae, Vespinae). Molecular Phylogenetics and Evolution, 107, 10-15.
0 Comments
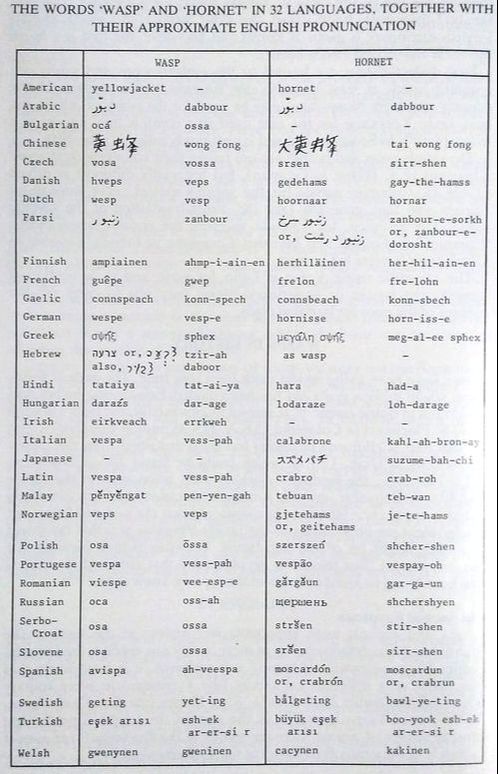
Do you love talking about wasps? Here's how you pronounce the word 'hornet' in dozens of languages!
References:
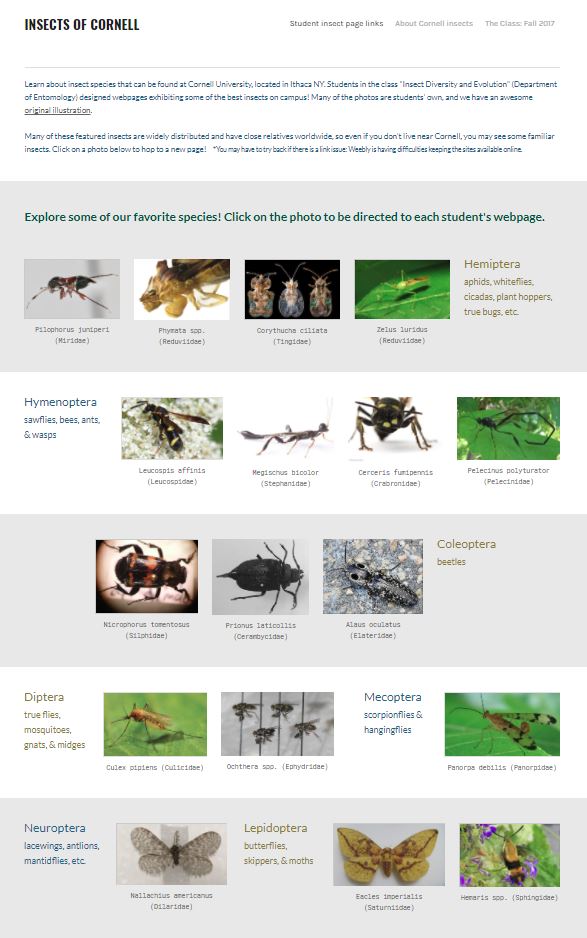
Edwards, R. (1980). Social wasps. Their biology and control. Rentokil Ltd. For the course I'm teaching this fall (Insect Diversity and Evolution, ENTOM 3310/3311), the students each designed a webpage on their favorite species of insect that could be found at Cornell. Check it out!
We used the free Weebly for Education platform. None of the students had made their own website prior to this semester & overall, it seemed like the students caught on quickly. The biggest problem I encountered is that the individual sites are often down. Clicking on the link gives a message: "This page isn't working" -- which is an issue on the Weebly end. That's a little disappointing, and I hope it improves.
Use the free photo processing program, Fiji, to make a gif out of your stack of z-stepped images! You'll produce a nice little video clip that scrolls through all of the layered shots that you've taken of your specimen.
make a gif using ImageJ and your stack of images:
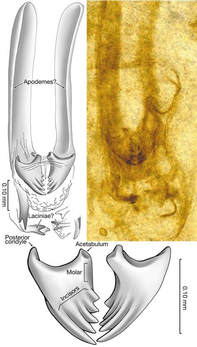
The fossil that was thought to be the oldest flying insect (and has been used to interpret and date the origins of flight) may not even be an insect at all! In an interesting turn, a recent publication asserts that the fossil Rhyniognatha hirsti is not the mandible of a flying insect, but is in fact a fragment of a myriapod: specifically, an immature centipede. In an article published May 30, 2017 in the journal PeerJ, Haug & Haug present evidence that the Devonian fossil R. hirsti (>400 My old) is not a flying insect... or an insect... or even closely related to insects... It is a centipede. The fossil is fragmentary, mainly mandibles and some other rather ambiguous parts connected to it. However, the shape of these mandibles was similar to those of dragonflies and neopterans, which prompted Engel and Grimaldi (2004) to tentatively assign it as a fragment of a flying insect. Now, Haug & Haug use 3D imaging and show remains of what they hypothesize is a myriapod-like head capsule. Also, the putative apodemes are deemed to be glands of ectodermal origin. The potential misplacement of this fossil has some pretty big implications. The next oldest fossils of flying insects are from the late Carboniferous -- about 80 My after this fossil! That is a huge time difference and affects our interpretation of the origins and evolution of insect flight. Also, many molecular phylogenetic studies have used this as a fossil calibration. For instance, the hexapod dated phylogeny of Misof et al. 2014 incorporated this fossil as a stem calibration for Dicondylia (winged insects + their sister group Diplura), with an age of 411.5 Mya. The reinterpretation of this fossil, if it is accepted, will have wide effects on future work on insect evolution and dating. As of September 23, there were ~1500 views of the article and no papers citing it, so we'll have to stay tuned for a response! references:
Engel, M.S. & Grimaldi, D. (2004) New light shed on the oldest insect. Nature 427, 627-630. Haug, C. & Haug, J.T. (2017) The presumed oldest flying insect: more likely a myriapod? PeerJ, 5, e3402. Misof, B., Liu, S., et al. (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science, 346, 763-767. |
PhyloBlogCovering topics of phylogenetics and systematics & other science-related news. Archives
October 2019
Categories
All
|


 RSS Feed
RSS Feed